Research
Cell division regulation in Staphyloccocus aureus
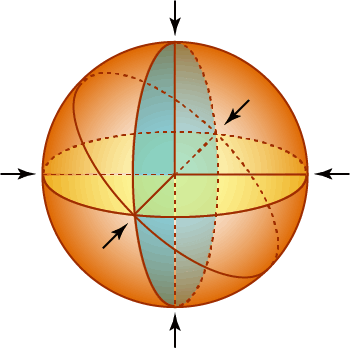
Staphylococcus aureus is a Gram-positive, versatile, opportunistic pathogen. Understandably, most S. aureus research has been aimed towards studying virulence factors, antibiotic-resistance mechanisms, and biofilm formation. Until recently, due to the lack of appropriate molecular tools and required technical advancement (for example, super-resolution microscopy, affordable deep-sequencing techniques, and CRISPR/Cas genetic engineering systems), studies focused on understanding basic biological processes such as cell division in this micron-sized organism has been very limited. Most rod-shaped bacteria divide in the center perpendicular to their long axis, therefore one could predict where the next division septum would appear in these organisms. How spherical bacteria choose their division site is an interesting fundamental biological question. In fact, S. aureus cells divide in sequential planes orthogonal to the preceding division planes. It has been known that S. aureus lacks a key division site determining system (Min system) that is present in both E. coli and B. subtilis. We plan to address these vast knowledge gaps in our understanding of cell division regulation in S. aureus by discovering factors that influence cell division and their mechanism of action. We have shown recently that in S. aureus, an essential protein, GpsB (which resembles another protein we study in the lab called DivIVA), is able to directly alter the polymerization kinetics of a tubulin-like key division protein, FtsZ (Eswara et al., eLife 2018). We also have discovered that YpsA, encoded by a gene immediately upstream of gpsB, is also involved in cell division in S. aureus (Brzozowski et al., Front. Microbiol. 2019).